This is the assignment of NTU CE7412: Computational and Systems Biology by Professor Jagath Rajapakse. The full report is here. It is the exercise of 5 interesting questions.
- Determine the entropy and the divergence of nucleotides and dinucleotides for Ecoli bacterial genome sequences.
- Model the sequence with (i) an independent model of sequences, (ii) a first-order Markov chain, and (iii) a second order Markov chain for the Worm genome.
- Given an aligned pair of sequences, determine whether a well-matched segment found by allowing two mismaches in the sequence pair is statistically significant or not.
- Model the CpG islands and nonCPG islands, determine a data-driven threshold for detecting a CpG island. Evaluate the sensitivity and specificity of your method on given CpG islands or non CpG islands.
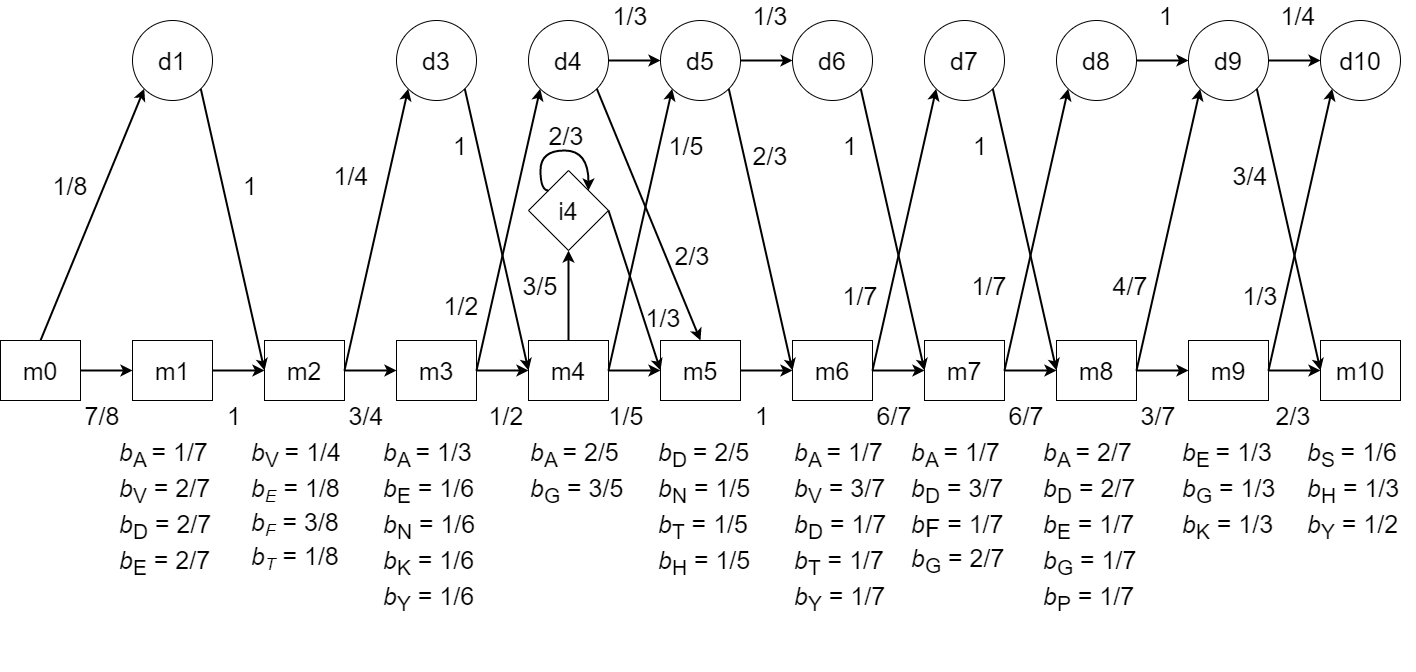
- Build a Profile HMM to represent the following multiple amino acid sequence alignment.