This is the assignment of NTU CE7412: Computational and Systems Biology by Professor Jagath Rajapakse. The full report is here.
This project aims to discover biomarkers and predict colorectal cancer (CRC) through studying the gene expressions of 59 CRC patients and 62 healthy controls (HCs) from Gene Expression Ominibus (GEO) database with id GSE164191. Two data objects are essential: a gene expression matrix (54,675, 121) and a phenotype table (121, 4). To achieve the goal, 4 types of analysis have been conducted as followings:
Differential Expression Analysis (DEGs)
I identify the significant differently expressed genes between CRC and HC groups with age and gender co-variants using a 0.05 p-value threshold. A DGEList object is created and normalized by the TMM method. As the library size ratio is smaller than 3-fold, counts are converted to logCPM for the limma-trend differential expression method. A linear model is fitted using Limma package.
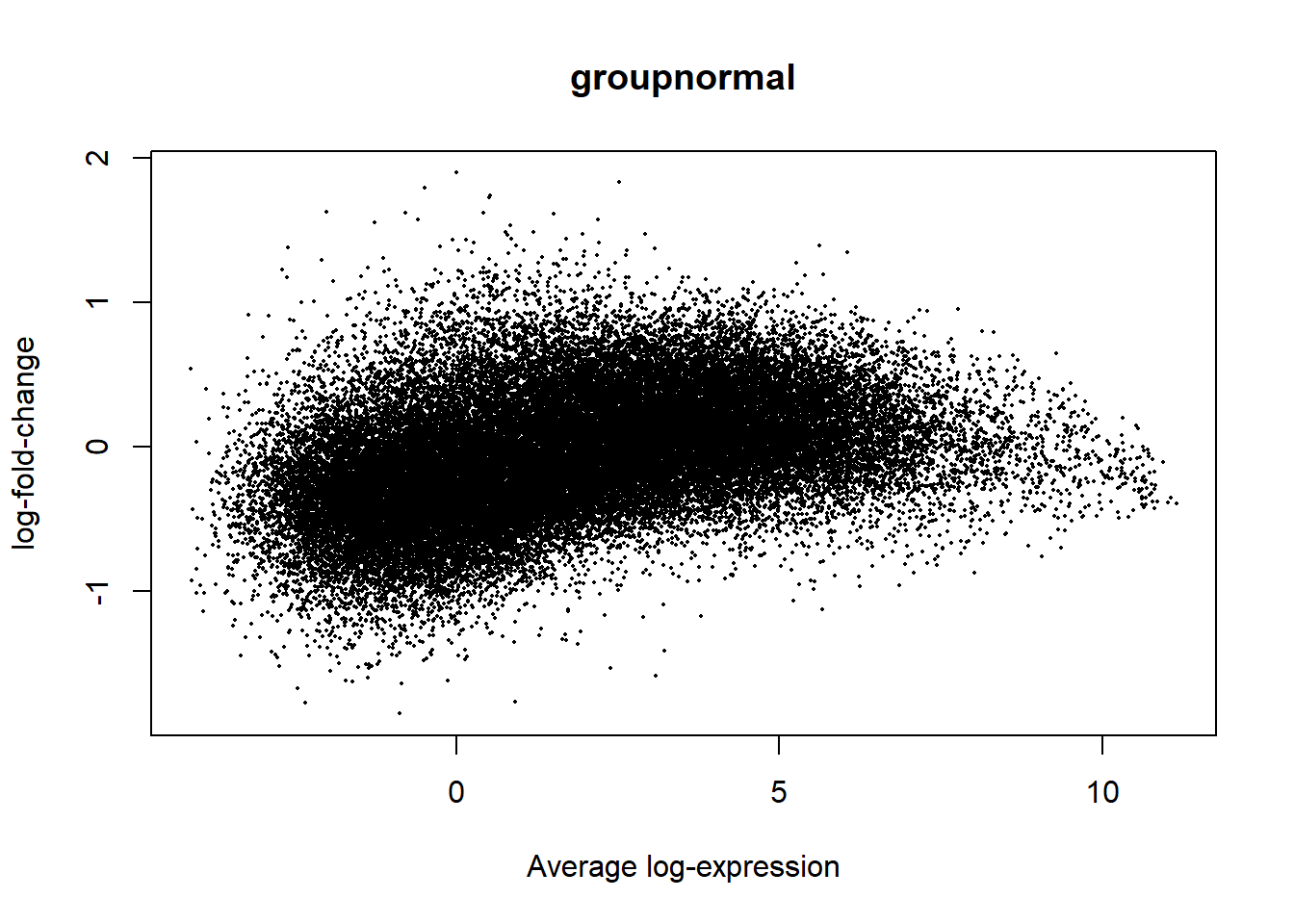 { width=50% }
{ width=50% }
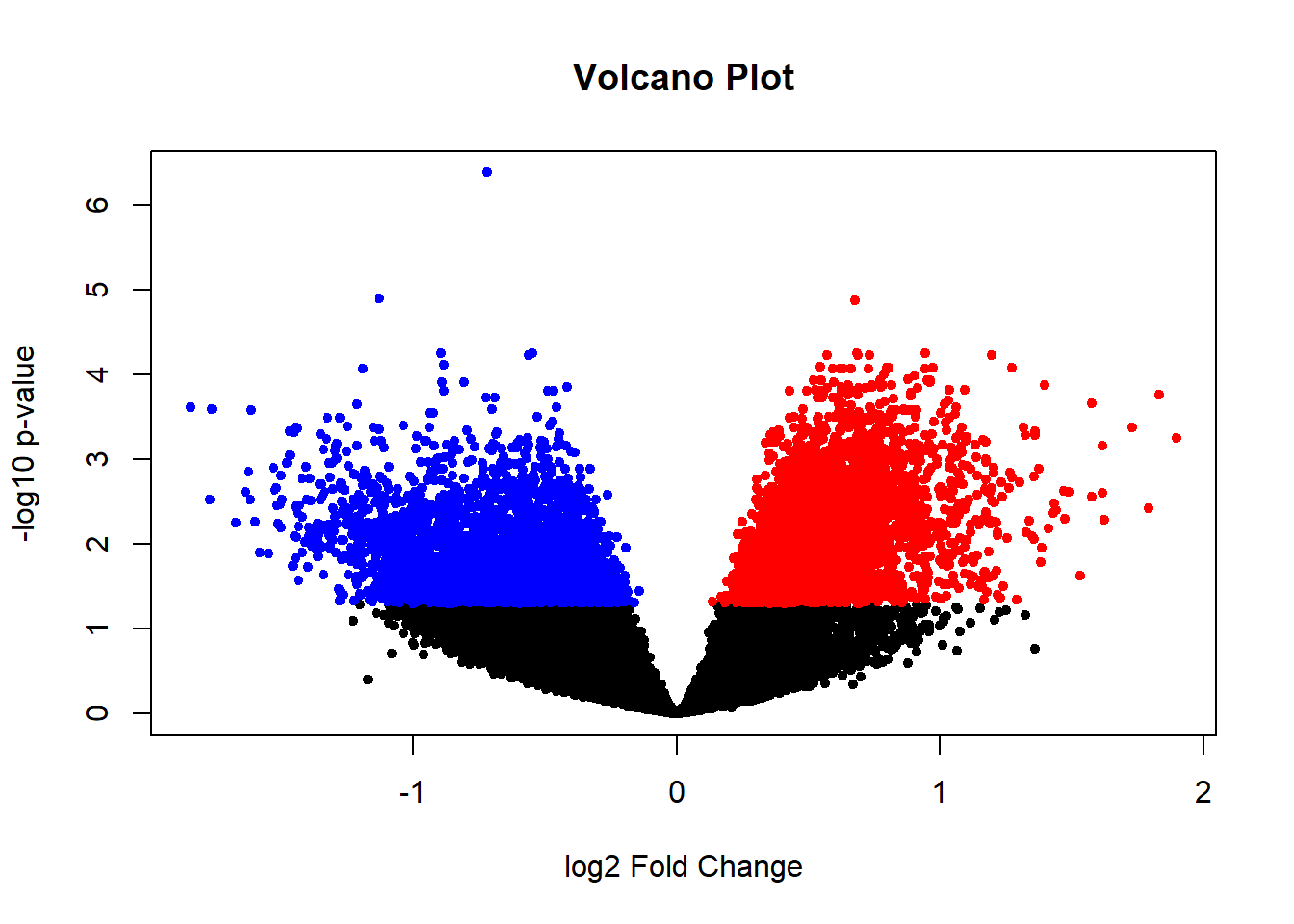 { width=50% }
{ width=50% }
Gene Ontology Enrichment Analysis (GOEA)
I identify the top enriched gene ontology terms and plot the GO hierarchy. A topGO object is created to perform GO enrichment test.
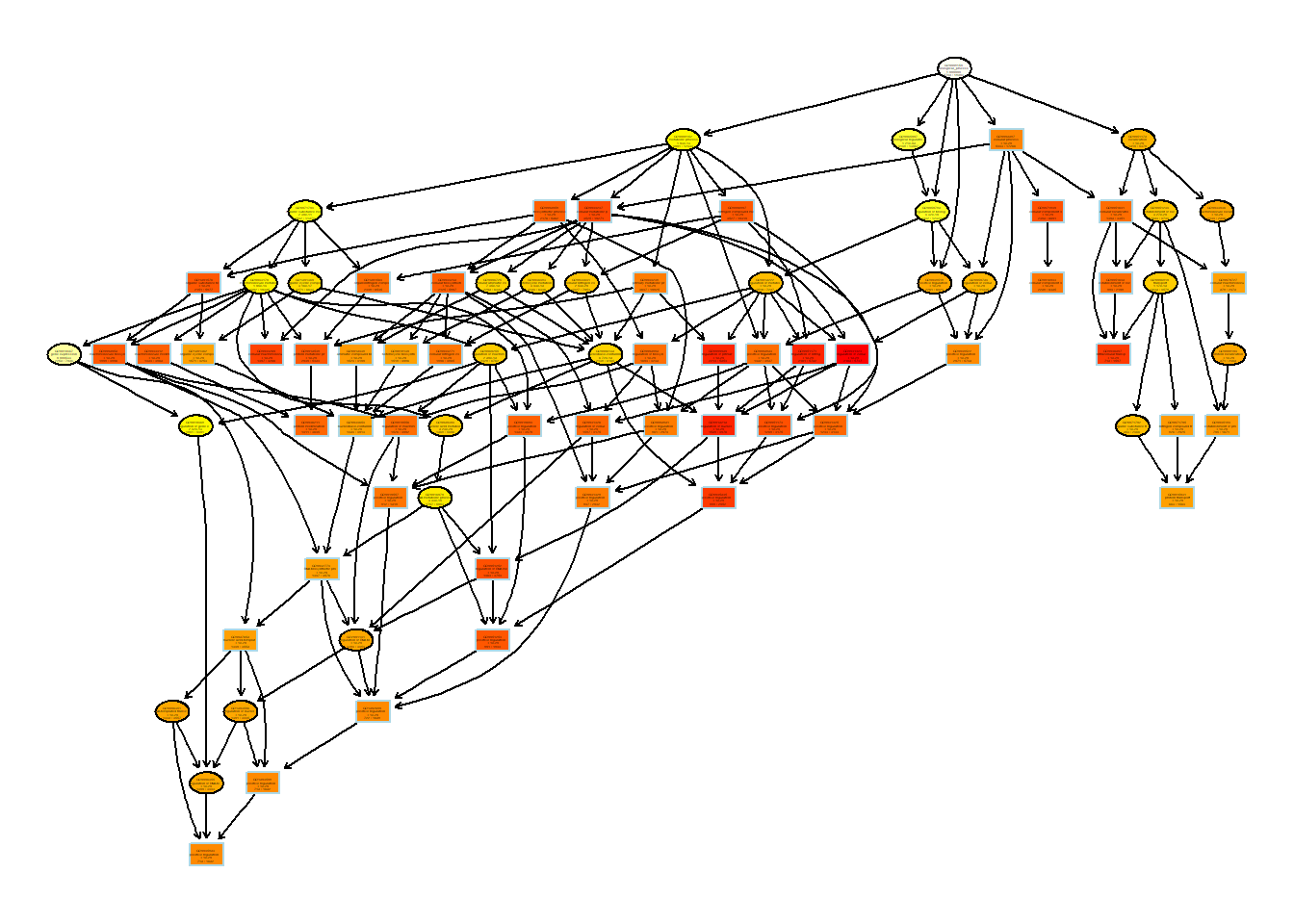 { width=50% }
{ width=50% }
Gene Set Enrichment Analysis (GSEA)
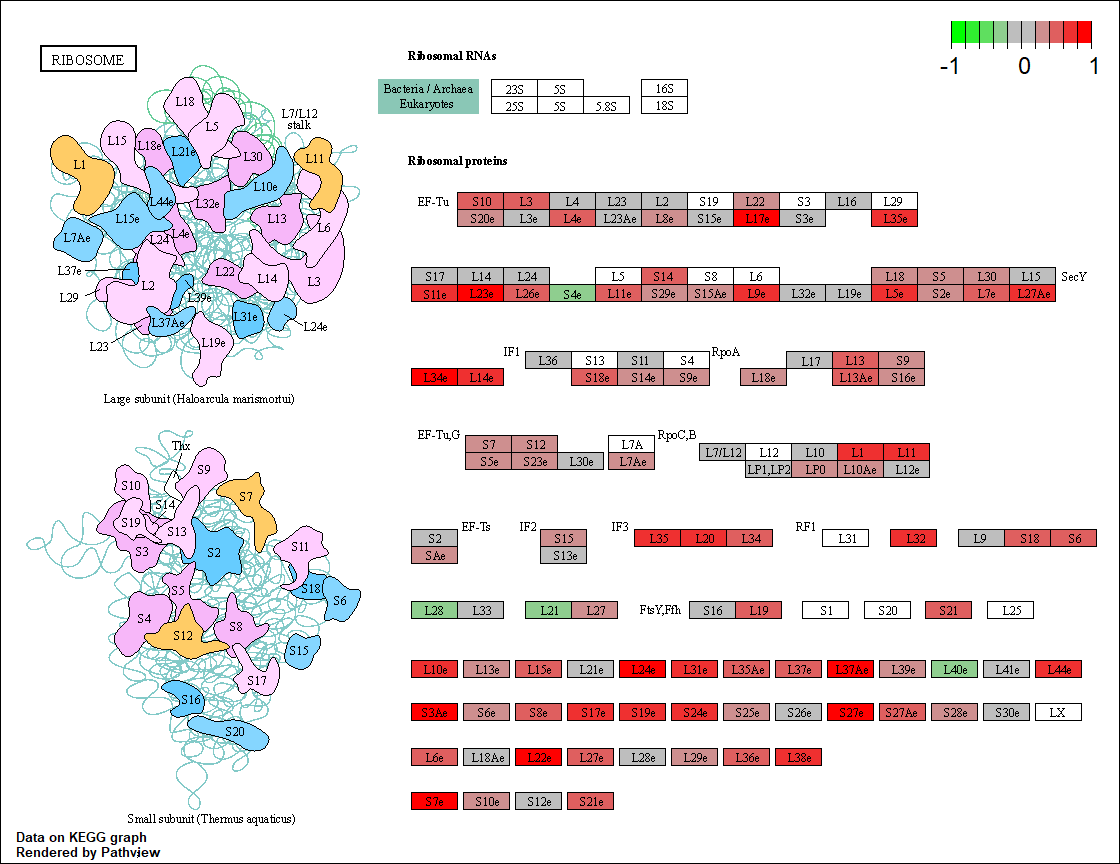
I identify the significantly enriched pathways against KEGG pathways with a 0.05 p-value threshold. The function gseKEGG from clusterProfiler is used to perform GSEA. The pathview library is used to plot two enriched pathways: hsa03010 and hsa03040. (to be updated)
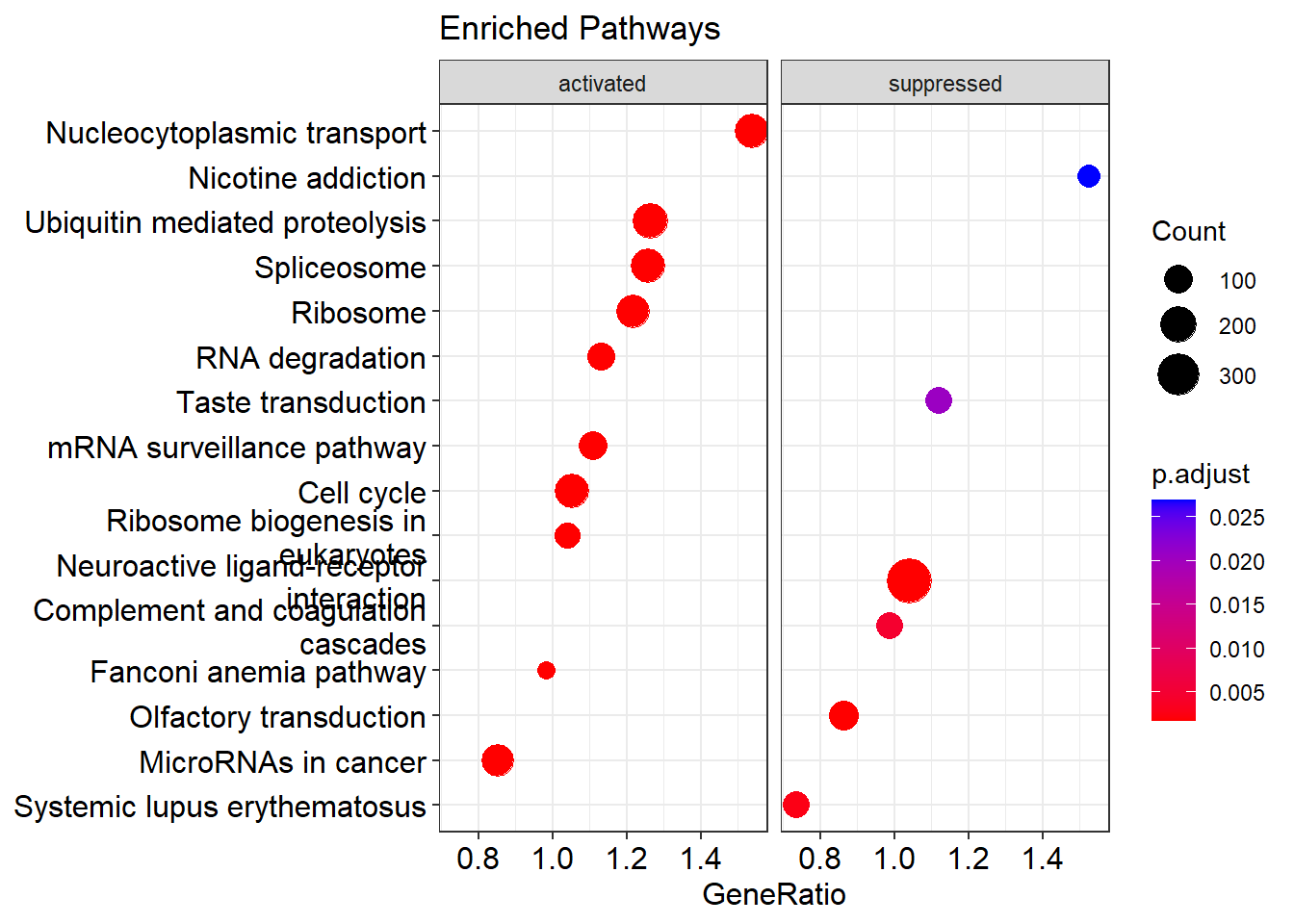 { width=50% }
{ width=50% }
 { width=50% }
{ width=50% }
CRC Prediction
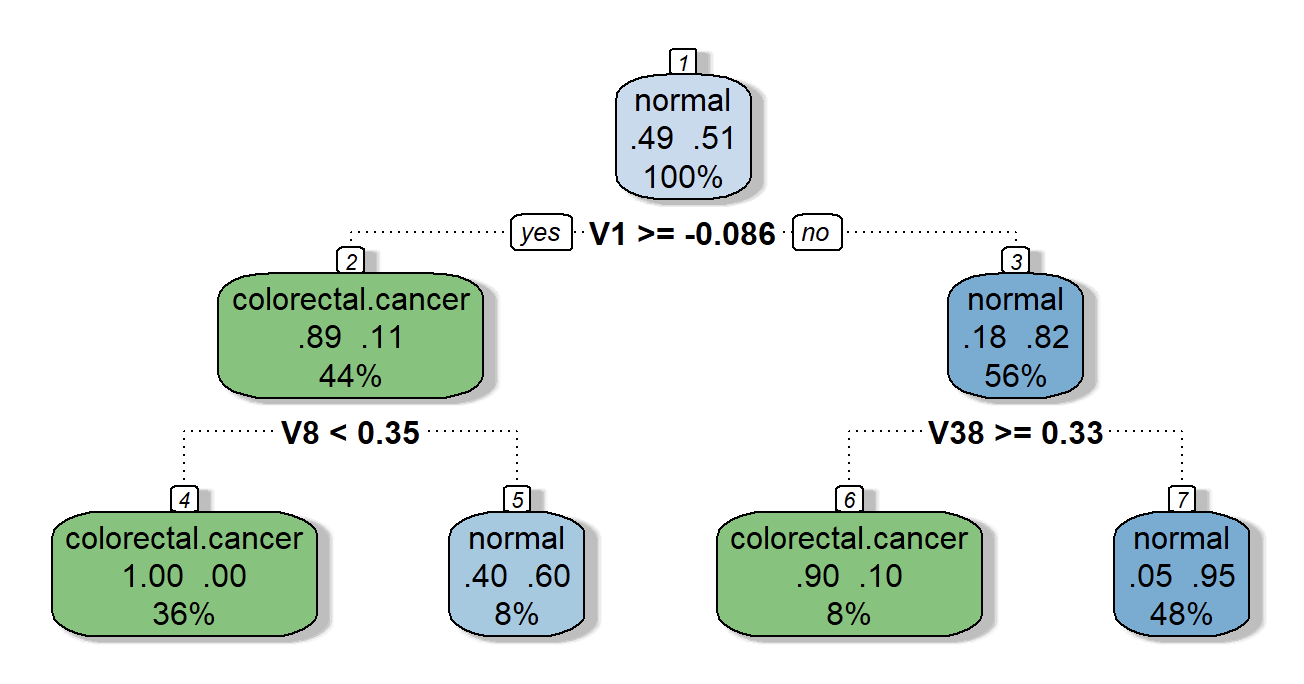
I build and tune the models to predict CRC patients and HCs based on the expressions of top 100 DEGs using caret library. Prediction models include linear discriminant analysis (LDA), generalized linear models (GLM), support vector machines (SVM), neural networks (NN), decision trees (DT), and gradient boost machines (GBM).
 { width=50% }
{ width=50% }
Through the above analysis, significant differences in gene expression are found between CRC patients and HCs, which could be effective biomarkers. Prediction models showed a high accuracy in this tiny datasets. Their real performance requires to be validated in larger datasets.
